1. Introduction
As we all know, forward neural networks (FNN) have no connection between neurons in the same layer or in cross layers. The propagation direction is single. Though it is easier to learn but to some extent, it reduces the power of neural networks.In FNN, the output only relies on the current input. However, in practice, the output is associated not only with the input of current epoch but past few epochs. Like, video, speech, text, those signals are hard to be handled by FNN.
Recurrent neural network (RNN) is one kind of NN that neurons can receive information from themselves and other neurons which forms a circuit. This model is more coherent to the neural structure of beings. RNN has been widely used in speech recognition, language models and natural language generation. So in this post, we are going dissect this model.
Neural networks, whether recurrent or feedforward, can receive as input raw sensor signals. However, applying them to features derived from the raw sensor signals often leads to higher performance. Discovering adequate features requires expert knowledge, which necessarily limits a systematic exploration of the feature space. Convolutional networks (CNNs) have been suggested to address this.
2. Background
2.1 Independent variables vs. Dependent variables
Before talking about RNN, we need to have an overview of the relationship between random variables. This relationship includes the independent form and the dependent form.
The independent variable is the variable whose change isn’t affected by any other variable in the experiment. In contrast, dependent variables describe what changes as a result of the changes of other dependent variables. As for multi independent variables, they are parallel. But they constitute a graphical structure for multi dependent variables, which is also called the probabilistic graphical model.
probabilistic graphical model
2.2 Probabilistic Graphical Model (PGM)
PGM is a probabilistic model which uses graphs to represent the relationship of dependent variables. A node in the graph can represent one or a group of variables. The side between two nodes shows the probabilistic relationship.
PGM can be divided into two classifications: undirected graph and directed graph. The directed graph is also called Bayesian Network (The Hidden Markov Model is the simplest dynamic Bayesian Model). The undirected graph is also called Markov Network.
2.3 Hidden Markov Model
In this model, the variables can be divided into two groups: status variables or hidden variables and observable variables.
Status variables or hidden variables cannot be observable. It can be used to give feedback to outside depending on different inputs.
observable variables are measurable that give clues to infer the status of a system.
3. Simple RNN model
3.1 Time Delay Neural Network (TDNN)
TDNN is a NN that add a time delay in non-output layers of feedforward neural network (FNN) to record a few recent outputs of a certain neuron. For example, X is a neuron in the input layer. Xt means the value that X sends to a neuron in the next layer at time eepoch t. So, Xt-1, Xt-2, ... Xt-p+1 means the recent p input of X. Then the function of this so-called time delay is to record those outputs {Xt, Xt-1, Xt-2, ... Xt-p+1} and generate a new input to the neuron in the next layer. That means, at epoch t, the neurons in l+1 layer are related to the recent p outputs of neurons in l layer.
 |
| Figure 3.1 |
If we consider the origin input as an image. The recent p inputs are like p pixels in a window. TDNN is like a simple convolutional neural network (CNN).
3.2 Nonlinear Autoregressive with Exogenous Inputs Model,NARX
Autoregressive Model (AR) is a model that is used to process temporal series, whose function is to predict the status of a variable depending on past status. It is employed to predict itself.
 |
| Figure 3.2 |
where, yt is the prediction of y at t epoch. w0 is a constant parameter and ε is the random noise (subject to normal distribution). w is the weight for each past status.
Nonlinear Autoregressive with Exogenous Inputs Model is a model that combines time delay NN and AR but it is nonlinear.
From the equation in figure 3.3, we can see that yt is related to recent p input x and output y.
f(·) means nonlinear function.
3.3 Simple Recurrent Network, SRN
For section 3.2, we apply it to neural networks, we can get a kind of classic neural network, recurrent neural network (RNN).
Given an input series X(1:T)=(X1.....XT). So the status of neurons in next hidden layer ht is expressed in the form:
 |
| Figure 3.4 |
h0=0. so we can give a RNN model like: (including input layer, hidden layer and output layer, each layer only has one neuron.)
 |
| Figure 3.5 |
According to feedforward neural network model with three layers, let zt stand the pure input of hidden layer. f(·) means the non-linear activation function, e.g. logistic, tanh and ReLU. W is the status-status (transformation) weight matrix, U is the input-status weight matrix, b is the bias. ht is the status at t. And yt means the output at t. V is the status-output weight matrix. Then we have:
So now we have a basic neural network unit for RNN. At each time epoch, the model will get an input. At the same time, the status will be transformed along the time. Since the main feature of RNN is the status transformation, it is not necessary to give an output at each epoch. So the output y is optional.
If we combine multi basic RNN models at different epochs and expanse in the order of time, we have:
 |
| Figure 3.6 |
If we combine multi basic RNN models at different epochs and expanse in the order of time, we have:
 |
| Figure 3.7 |
 |
| Figure 3.7-2 |
3.4 Stacked Recurrent Neural Network,SRNN
The power of neural networks relies on the depth of networks. More hidden layers, stronger learning ability neural networks are.
Based on section 3.3, we increase the depth of hidden layers, then we have:
So the status equation turns:
From the figure 3.8 and 3.9, we can see the link weight between 2 layers at the same epoch is the input-status matrix, U; the link weight between 2 status in the same layer is the status-status matrix, W; Each hidden layer neuron has a bias, b. The so-called sharing weights are those parameters in the same color are same.
This is how to stack multi hidden layers for RNN models. Until now, what I show you is the RNN with a very simple structure whose feature is T input, T status in each hidden layer and T output. Actually, RNN can be adapted to different structures to apply to different problems.
 |
| Figure 3.8 |
 |
| Figure 3.9 |
This is how to stack multi hidden layers for RNN models. Until now, what I show you is the RNN with a very simple structure whose feature is T input, T status in each hidden layer and T output. Actually, RNN can be adapted to different structures to apply to different problems.
4. Three different modalities of RNN
RNN can be applied to many different forms of machine learning tasks. There are mainly three modalities of RNN: Sequence to Classification model, Synchronous Sequence to Sequence (Seq2Seq) model and asynchronous Sequence to Sequence model (also called Encoder-Decoder).
4.1 Sequence to Classification model (Seq2C)
Seq2C is mainly used to classify sequential data. The input is a sequence and the output is classification. For instance, in text classification, the input is sequential words and the output is the classification of this text.
Assume an input X(1:T)=(X1.....XT). The length is T. The output y is {1, ..., C}. We can input this X at different time epoch, like {X1, X2, X3, ..., XT) into RNN. The status we can get is {h1, h2, h3, ..., hT}. Instead of giving an output at each epoch, we only give one output for classification.
Here we have 2 forms. One form is to take the hT as the final feature and give it to a classifier. The other is take the mean of the whole status as the final feature and send it to a classifier.
Similarly, we assume the input X is X(1:T)=(X1.....XT). The output is Y(1:M)=(y1, ..., yM). The process is realized by encoding and then decoding as shown in the figure below:
Under supervised learning, given an training example, (x,y): X(1:T)=(X1.....XT); Y(1:T)=(y1, ..., yT). That is to say, at each epoch, there is an output yt. Then the loss function (e.g. cross entropy and mean square error) at epoch t is:
As I said in a post about gradient descent and random gradinet descent, we often use batch
gradient descent to train parameters. So the loss function is the sum of loss functions of a batch of examples. So,
The inside of RNN has a recurrent structure, so the learning style is a little different from the way of feedforward neural networks. There are two ways to compute the gradient: Backpropagation through time (BPTT); Real-time recurrent learning (RTRL).
Then we need to compute the partial derivation of the sum loss function subject to the parameters. Combining the matrix in figure 5.2-1, we have:
Here, i and j stand the row and column of the matrix W, respectively. [hk-1]j is the jth component of the status at k-1 epoch. IIi(x) is the short for the vector [0,0,...[hk-1]j,...,0]T
Because there is a recursive relationship between statuses. We assume δ(t,k) means the derivative of the loss at t epoch subject to the pure input zk at k epoch, then:
Therefore, in order to compute the gradient, we need a whole feedforward computation and back computation to update the parameters.
Unlike BPTT, RTRL computes the gradient via feedforward style.
Comparison:
BPTT and RTRL are all based on gradient descent, feedforward model and back model apply chain rules to compute gradient. In RNN, the dimension of output is far smaller than the dimension of inputs. Thus, the computation load of BTPP is low but it needs to store intermediate gradients at all epochs which increase the space complexity. RTRL does not need feedback of gradients. Therefore, it is very suitable for online learning or infinite sequence learning.
In order to solve this problem, Researchers invented the Gated RNN.
Let's look at the structure of a LSTM recurrent unit. we can see all input (including observations and outer status at last epoch) will be sent to three gates and the inner status, respectively. Then the activation function determines how these gates and inner status work (sounds like natural selection).
https://keras.io/
Assume an input X(1:T)=(X1.....XT). The length is T. The output y is {1, ..., C}. We can input this X at different time epoch, like {X1, X2, X3, ..., XT) into RNN. The status we can get is {h1, h2, h3, ..., hT}. Instead of giving an output at each epoch, we only give one output for classification.
Here we have 2 forms. One form is to take the hT as the final feature and give it to a classifier. The other is take the mean of the whole status as the final feature and send it to a classifier.
 |
| Figure 4.1 |
4.2 Synchronous Sequence to Sequence
Synchronous Sequence to Sequence model is mainly used for sequence labeling. That is to say, there is an output at each epoch which I showed you in section 3.3. |
| Figure 4.2 |
4.3 Asynchronous Sequence to Sequence (Encoder-Decoder)
Asynchronous Sequence to Sequence is also called Encoder-Decoder model which has been used widely. This model doesn't require the strict reflection relationship between the input sequence and the output sequence as well as the same length. For instance, the input is word sequence, the output is the word sequence of a target language.Similarly, we assume the input X is X(1:T)=(X1.....XT). The output is Y(1:M)=(y1, ..., yM). The process is realized by encoding and then decoding as shown in the figure below:
 |
| Figure 4.3 |
 |
| Figure 4.4 Encoder-Decoder model example |
5. Parameters learning of RNN
Basically, the parameters of RNN also learn via gradient descent.Under supervised learning, given an training example, (x,y): X(1:T)=(X1.....XT); Y(1:T)=(y1, ..., yT). That is to say, at each epoch, there is an output yt. Then the loss function (e.g. cross entropy and mean square error) at epoch t is:
 |
| Figure 5.1 |
gradient descent to train parameters. So the loss function is the sum of loss functions of a batch of examples. So,
 |
| Figure 5.2 |
5.2 The number of parameters
From Figure 3.6, we can see all parameters we have are U, W, b and V(optional). So If we only consider parameters in the hidden layers, U, W and b are included.
The input is current xt and previous state ht-1. The dimension of x and h keep unchanged. If the size of x is m and h is n. The sum of parameters (U,W,b) is (m+n)*n+n.
Thus, the status-status weight is a n*n matrix.
The input-status weight is a m*n matrix.
The bias is 1*n.
When use matrix, the relationship can be expressed in the form:
 |
| Figure 5.2-1 |
5.2 Backpropagation through time (BPTT)
The feature of BPTT is that the loss function at epoch t only propagates back to the parameters before t epoch (inclusive). Like shown below, the yellow arrow only relates the parameters at t-2 as well as before t-2. And the blue arrow and the red arrow all follow this rule.
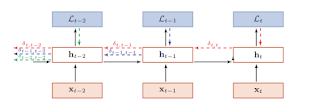 |
| Figure 5.3 BPTT |
Considering Figure 3.6, we have:
 |
| Figure 5.4 |
Here an important feature of RNN is that the basic RNN model shares all parameters (including U, W and b) after expansion along time. For Stacked Recurrent Neural Networks (shown in Figure 3.8), the parameters in the same layer are same and in different layers are different. Here is an example:
 |
| Figure 5.5 |
 |
| Figure 5.6 |
Because there is a recursive relationship between statuses. We assume δ(t,k) means the derivative of the loss at t epoch subject to the pure input zk at k epoch, then:
 |
| Figure 5.7 |
 |
| Figure 5.8 |
5.3 Real-time recurrent learning (RTRL)
The purpose of RTRL is to give an output at each epoch. If at t+1, we give an output and get a loss function. Then we compute the derivative of the current loss function: |
| Figure 5.9 |
Comparison:
BPTT and RTRL are all based on gradient descent, feedforward model and back model apply chain rules to compute gradient. In RNN, the dimension of output is far smaller than the dimension of inputs. Thus, the computation load of BTPP is low but it needs to store intermediate gradients at all epochs which increase the space complexity. RTRL does not need feedback of gradients. Therefore, it is very suitable for online learning or infinite sequence learning.
6. Gated RNN
RNN face two problems, gradient exploding problem and gradient vanishing problem as shown below. In theory, RNN can build the relationship between current status and all past statuses. However, when the time interval is longer enough, RNN will appear these two problems leading to RNN only can learn short-term relationship. The model determines the long dependencies of RNN. This is so-called Long-Term Dependencies Problem. |
| Figure 6.1 |
In order to solve this problem, Researchers invented the Gated RNN.
6.1 Long short-term memory, LSTM
LSTM is the most successful RNN model that has been widely applied in many areas, e.g. speech recognition, machine interpretation, voice model and text generation. LSTM mitigates the long-term dependencies problem via linear connections. Linear connections and gated control are very effective to avoid the gradient vanishing problem. In fact, this mechanism also can be applied to deep feedforward neural networks.
In essence, adding linear connections for variables is more close to the way things happen. We know that things are always related to each other. Considering a more complex relationship can recover more potential relations which are not easy to understand. Graph is a good model to establish more connections between things. So it is easy to say that RNN is kind of a graph network. The graph network is a emerging research direction which is not mature.
Then, let's have a look at the structure inside of LSTM.
From figure 6.2, we can see apart from the input and status, some other variables are included.
Three kinds of gates:
When ft =0, it=1, memory unit will be cleared out. ct=c't.
When ft =1, it=0, memory unit will copy the previous content. ct=ct-1.
In essence, adding linear connections for variables is more close to the way things happen. We know that things are always related to each other. Considering a more complex relationship can recover more potential relations which are not easy to understand. Graph is a good model to establish more connections between things. So it is easy to say that RNN is kind of a graph network. The graph network is a emerging research direction which is not mature.
Then, let's have a look at the structure inside of LSTM.
From figure 6.2, we can see apart from the input and status, some other variables are included.
 |
| Figure 6.2 |
- forget gate: ft, control how much information should the inner status at last epoch, ct-1, forget.
- input gate: it, control how much information should the candidate inner status c't save.
- output gate: ot control how much information should the real inner status ct send out to outer status ht.
When ft =0, it=1, memory unit will be cleared out. ct=c't.
When ft =1, it=0, memory unit will copy the previous content. ct=ct-1.
| Figure 6.3 LSTM recurrent unit |
 |
| Figure 6.4 |
6.2 Gated Recurrent Unit,GRU
Reference
https://zhuanlan.zhihu.com/p/27901936
https://zhuanlan.zhihu.com/p/38816145
https://nndl.github.io/chap-%E5%BE%AA%E7%8E%AF%E7%A5%9E%E7%BB%8F%E7%BD%91%E7%BB%9C.pdf




























